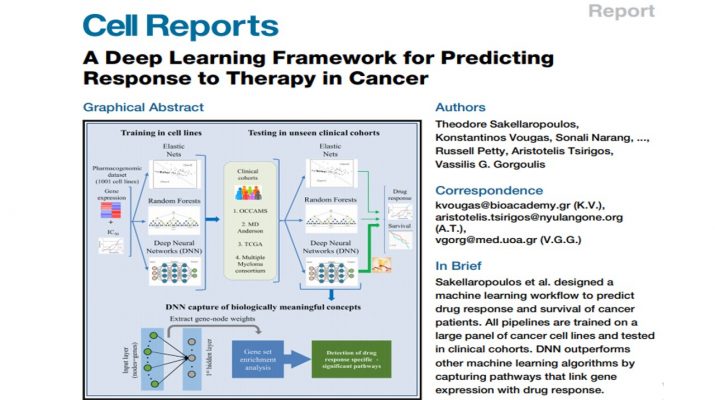
Sakellaropoulos et al. designed a machine learning workflow to predict drug response and survival of cancer patients. All pipelines are trained on a large panel of cancer cell lines and tested in clinical cohorts. DNN outperforms other machine learning algorithms by capturing pathways that link gene expression with drug response.
Predicting the clinical response to therapeutic agents is a major challenge in cancer treatment. To deliver personalized treatment with high efficacy, identifying molecular disease sig-natures and matching them with the most effective therapeutic interventions are essential. The advent of the ‘‘omics’’ era has permitted scientists to dissect the molecular events that are known to drive carcinogenesis. Nonetheless, effective translation of the growing wealth of high-throughput profiling data into clinically meaningful results has been challenging.
Summary
A major challenge in cancer treatment is predicting clinical response to anti-cancer drugs on a personalized basis. Using a pharmacogenomics database of1,001 cancer cell lines, we trained deep neural networks for prediction of drug response and assessed their performance on multiple clinical cohorts. We demonstrate that deep neural networks outperform the current state in machine learning frameworks. We provide a proof of concept for the use of deep neural network-based frameworks to aid precision oncology strategies.
Highlights
* A machine learning (ML) workflow is designed to predict drug response in cancer patients
* Deep neural networks (DNNs) surpass current ML algorithms in drug response prediction
* DNNs predict drug response and survival in various large clinical cohorts
* DNNs capture intricate biological interactions linked to specific drug response pathways
Authors
Theodore Sakellaropoulos,
Konstantinos Vougas, Sonali Narang, …,
Russell Petty, Aristotelis Tsirigos,
Vassilis G. Gorgoulis
Source: https://www.cell.com/cell-reports/pdf/S2211-1247(19)31488-3.pdf