MR Fingerprinting (MRF) is a relatively new imaging framework for MR acquisition, reconstruction, and analysis, which can provide rapid, efficient and simultaneous quantification of multiple tissue properties from a single acquisition. Compared to conventional MR imaging approaches, MRF uses pseudorandomized acquisition parameters (such as flip angles and repetition times) to generate unique signal signatures for different tissue types and retrieve quantitative tissue parameters using a template matching algorithm.
Since its introduction in 2013, this technique has been successfully applied for quantitative imaging of multiple human organs including the brain, abdominal organs, heart, and breast. The quantitative tissue properties obtained using MRF, such as T1 and T2 relaxation times, have been demonstrated to provide new insights into improved tissue characterization and disease diagnosis. Specifically for brain imaging, MRF has been applied for longitudinal characterization of brain development in early childhood, differentiation of brain tumor types, and improved detection and diagnosis of epileptic lesions.
Abstract
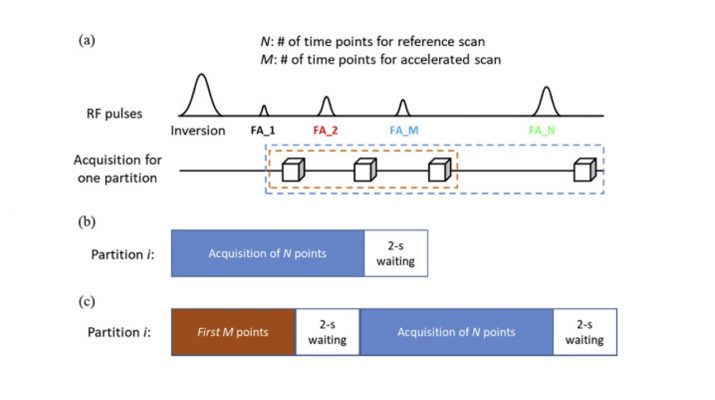
MR Fingerprinting (MRF) is a relatively new imaging framework capable of providing accurate and simultaneous quantification of multiple tissue properties for improved tissue characterization and disease diagnosis. While 2D MRF has been widely available, extending the method to 3D MRF has been an actively pursued area of research as a 3D approach can provide a higher spatial resolution and better tissue characterization with an inherently higher signal-to-noise ratio. However, 3D MRF with a high spatial resolution requires lengthy acquisition times, especially for a large volume, making it impractical for most clinical applications.
In this study, a high-resolution 3D MR Fingerprinting technique, combining parallel imaging and deep learning, was developed for rapid and simultaneous quantification of T1 and T2 relaxation times. Parallel imaging was first applied along the partition-encoding direction to reduce the amount of acquired data. An advanced convolutional neural network was then integrated with the MRF framework to extract features from the MRF signal evolution for improved tissue characterization and accelerated mapping. A modified 3D-MRF sequence was also developed in the study to acquire data to train the deep learning model that can be directly applied to prospectively accelerate 3D MRF scans. Our results of quantitative T1 and T2 maps demonstrate that improved tissue characterization can be achieved using the proposed method as compared to prior methods.
With the integration of parallel imaging and deep learning techniques, whole-brain (26 × 26 × 18 cm3) quantitative T1 and T2 mapping with 1-mm isotropic resolution were achieved in ~7 min. In addition, a ~7-fold improvement in processing time to extract tissue properties was also accomplished with the deep learning approach as compared to the standard template matching method. All of these improvements make high-resolution whole-brain quantitative MR imaging feasible for clinical applications.
Highlights
- 3D MRF with 1-mm isotropic resolution and whole-brain coverage for neuroimaging.
- Integration of advanced parallel imaging and deep learning techniques for accelerated quantitative imaging.
- Whole-brain (26 × 26 × 18 cm3) T1 and T2 mapping in ~7 min.
- 7-fold improvement in processing time to extract tissue properties as compared to the standard template matching approach.
Yong Chen
Zhenghan Fang
Sheng-Che Hun
Wei-Tang Chang
Dinggang Shen
Weili Lin